Page 181 - Contributed Paper Session (CPS) - Volume 2
P. 181
CPS1496 Tim Christopher D.L et al.
(conditional on the inclusion of predictions from other machine learning
models).
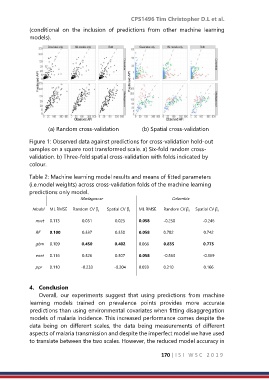
(a) Random cross-validation (b) Spatial cross-validation
Figure 1: Observed data against predictions for cross-validation hold-out
samples on a square root transformed scale. a) Six-fold random cross-
validation. b) Three-fold spatial cross-validation with folds indicated by
colour.
Table 2: Machine learning model results and means of fitted parameters
(i.e.model weights) across cross-validation folds of the machine learning
predictions only model.
Madagascar Colombia
Model ML RMSE Random CV β ̅ Spatial CV β ̅ i ML RMSE Random CV β ̅ i Spatial CV β ̅
i
i
nnet 0.113 0.031 0.025 0.058 -0.250 -0.246
RF 0.100 0.337 0.350 0.058 0.782 0.742
gbm 0.109 0.450 0.402 0.066 0.835 0.775
enet 0.116 0.326 0.307 0.058 -0.563 -0.369
ppr 0.110 -0.233 -0.204 0.059 0.210 0.166
4. Conclusion
Overall, our experiments suggest that using predictions from machine
learning models trained on prevalence points provides more accurate
predictions than using environmental covariates when fitting disaggregation
models of malaria incidence. This increased performance comes despite the
data being on different scales, the data being measurements of different
aspects of malaria transmission and despite the imperfect model we have used
to translate between the two scales. However, the reduced model accuracy in
170 | I S I W S C 2 0 1 9